At surface level - sorting a table may seem easy, and in many cases it is. But in a handful of cases it can get quite tricky, with some odd situations that need to be handled carefully. For this reason, we found it necessary to dedicate an entire vignette to just sorting and handling columns output by Tplyr.
Let’s start by looking at an example.
t <- tplyr_table(tplyr_adsl, TRT01A) %>%
add_total_group() %>%
add_treat_grps(Treated = c("Xanomeline Low Dose", "Xanomeline High Dose")) %>%
add_layer(
group_count(EOSSTT, by = SEX)
) %>%
add_layer(
group_desc(HEIGHTBL, by = SEX)
) %>%
build()
kable(t)| row_label1 | row_label2 | var1_Placebo | var1_Total | var1_Treated | var1_Xanomeline High Dose | var1_Xanomeline Low Dose | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|---|---|---|---|---|
| F | COMPLETED | 34 ( 39.5%) | 64 ( 25.2%) | 30 ( 17.9%) | 13 ( 15.5%) | 17 ( 20.2%) | 1 | 1 | 1 |
| F | DISCONTINUED | 19 ( 22.1%) | 79 ( 31.1%) | 60 ( 35.7%) | 27 ( 32.1%) | 33 ( 39.3%) | 1 | 1 | 2 |
| M | COMPLETED | 24 ( 27.9%) | 46 ( 18.1%) | 22 ( 13.1%) | 14 ( 16.7%) | 8 ( 9.5%) | 1 | 2 | 1 |
| M | DISCONTINUED | 9 ( 10.5%) | 65 ( 25.6%) | 56 ( 33.3%) | 30 ( 35.7%) | 26 ( 31.0%) | 1 | 2 | 2 |
| F | n | 53 | 143 | 90 | 40 | 50 | 2 | 1 | 1 |
| F | Mean (SD) | 156.06 ( 8.010) | 157.25 ( 7.374) | 157.94 ( 6.924) | 158.02 ( 6.370) | 157.88 ( 7.401) | 2 | 1 | 2 |
| F | Median | 156.20 | 157.50 | 157.50 | 157.50 | 157.85 | 2 | 1 | 3 |
| F | Q1, Q3 | 149.90, 162.60 | 152.40, 162.60 | 154.00, 162.60 | 154.28, 164.12 | 154.00, 162.60 | 2 | 1 | 4 |
| F | Min, Max | 137.2, 174.0 | 135.9, 175.3 | 135.9, 175.3 | 146.1, 170.2 | 135.9, 175.3 | 2 | 1 | 5 |
| F | Missing | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 6 |
| M | n | 33 | 111 | 78 | 44 | 34 | 2 | 2 | 1 |
| M | Mean (SD) | 173.03 ( 8.088) | 172.55 ( 7.946) | 172.34 ( 7.929) | 172.91 ( 7.304) | 171.60 ( 8.729) | 2 | 2 | 2 |
| M | Median | 174.00 | 172.70 | 172.70 | 172.70 | 172.10 | 2 | 2 | 3 |
| M | Q1, Q3 | 170.20, 177.80 | 168.25, 177.80 | 167.60, 177.80 | 170.15, 177.80 | 165.42, 177.48 | 2 | 2 | 4 |
| M | Min, Max | 144.8, 185.4 | 144.8, 195.6 | 147.3, 195.6 | 147.3, 190.5 | 157.5, 195.6 | 2 | 2 | 5 |
| M | Missing | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 6 |
In this table, we have:
- Added a ‘Total’ treatment group
- Added a ‘Treated’ group made up of the two treated cohorts
- Created a count layer for End of Study Status, reported by sex
- Created a descriptive statistics layer for Height at Baseline, reported by sex
Now let’s dig in.
Sorting Table Columns
Ordering Helpers
Ordering helpers are columns added into Tplyr tables to make sure that you can sort the display to your preference. In general, Tplyr will create:
- One order variable to order layers
- One order variable for each by variable
- At least one order variable for the target variables
In the example above, the t table outputs with three
columns:
-
ord_layer_indexindexes the layer itself. -
ord_layer_1indexes the first by variable,SEX. No options were presented so sorting was done alphabetically. -
ord_layer_2indexes the values of theEOSSTTvariable in the count layer, and the names of the summaries in the desc layer.
t %>%
select(starts_with("ord")) %>%
kable()| ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|
| 1 | 1 | 1 |
| 1 | 1 | 2 |
| 1 | 2 | 1 |
| 1 | 2 | 2 |
| 2 | 1 | 1 |
| 2 | 1 | 2 |
| 2 | 1 | 3 |
| 2 | 1 | 4 |
| 2 | 1 | 5 |
| 2 | 1 | 6 |
| 2 | 2 | 1 |
| 2 | 2 | 2 |
| 2 | 2 | 3 |
| 2 | 2 | 4 |
| 2 | 2 | 5 |
| 2 | 2 | 6 |
Reordering and Dropping Columns
Column selection from data frames is something that is already very
well done in R. The functions dplyr::select(),
magrittr::extract(), and [ can all be used to
reorder and drop column cleanly and concisely based on a user’s
preference.
To drop the ordering helpers, you can easily subtract them with ‘dplyr’ and ‘tidyselect’.
t %>%
select(-starts_with("ord_")) %>%
kable()| row_label1 | row_label2 | var1_Placebo | var1_Total | var1_Treated | var1_Xanomeline High Dose | var1_Xanomeline Low Dose |
|---|---|---|---|---|---|---|
| F | COMPLETED | 34 ( 39.5%) | 64 ( 25.2%) | 30 ( 17.9%) | 13 ( 15.5%) | 17 ( 20.2%) |
| F | DISCONTINUED | 19 ( 22.1%) | 79 ( 31.1%) | 60 ( 35.7%) | 27 ( 32.1%) | 33 ( 39.3%) |
| M | COMPLETED | 24 ( 27.9%) | 46 ( 18.1%) | 22 ( 13.1%) | 14 ( 16.7%) | 8 ( 9.5%) |
| M | DISCONTINUED | 9 ( 10.5%) | 65 ( 25.6%) | 56 ( 33.3%) | 30 ( 35.7%) | 26 ( 31.0%) |
| F | n | 53 | 143 | 90 | 40 | 50 |
| F | Mean (SD) | 156.06 ( 8.010) | 157.25 ( 7.374) | 157.94 ( 6.924) | 158.02 ( 6.370) | 157.88 ( 7.401) |
| F | Median | 156.20 | 157.50 | 157.50 | 157.50 | 157.85 |
| F | Q1, Q3 | 149.90, 162.60 | 152.40, 162.60 | 154.00, 162.60 | 154.28, 164.12 | 154.00, 162.60 |
| F | Min, Max | 137.2, 174.0 | 135.9, 175.3 | 135.9, 175.3 | 146.1, 170.2 | 135.9, 175.3 |
| F | Missing | 0 | 0 | 0 | 0 | 0 |
| M | n | 33 | 111 | 78 | 44 | 34 |
| M | Mean (SD) | 173.03 ( 8.088) | 172.55 ( 7.946) | 172.34 ( 7.929) | 172.91 ( 7.304) | 171.60 ( 8.729) |
| M | Median | 174.00 | 172.70 | 172.70 | 172.70 | 172.10 |
| M | Q1, Q3 | 170.20, 177.80 | 168.25, 177.80 | 167.60, 177.80 | 170.15, 177.80 | 165.42, 177.48 |
| M | Min, Max | 144.8, 185.4 | 144.8, 195.6 | 147.3, 195.6 | 147.3, 190.5 | 157.5, 195.6 |
| M | Missing | 0 | 0 | 0 | 0 | 0 |
Or you can reorder columns. In this example the “Total” result column is moved to the front of the results.
t %>%
select( starts_with("row"), var1_Total, starts_with("var1")) %>%
kable()| row_label1 | row_label2 | var1_Total | var1_Placebo | var1_Treated | var1_Xanomeline High Dose | var1_Xanomeline Low Dose |
|---|---|---|---|---|---|---|
| F | COMPLETED | 64 ( 25.2%) | 34 ( 39.5%) | 30 ( 17.9%) | 13 ( 15.5%) | 17 ( 20.2%) |
| F | DISCONTINUED | 79 ( 31.1%) | 19 ( 22.1%) | 60 ( 35.7%) | 27 ( 32.1%) | 33 ( 39.3%) |
| M | COMPLETED | 46 ( 18.1%) | 24 ( 27.9%) | 22 ( 13.1%) | 14 ( 16.7%) | 8 ( 9.5%) |
| M | DISCONTINUED | 65 ( 25.6%) | 9 ( 10.5%) | 56 ( 33.3%) | 30 ( 35.7%) | 26 ( 31.0%) |
| F | n | 143 | 53 | 90 | 40 | 50 |
| F | Mean (SD) | 157.25 ( 7.374) | 156.06 ( 8.010) | 157.94 ( 6.924) | 158.02 ( 6.370) | 157.88 ( 7.401) |
| F | Median | 157.50 | 156.20 | 157.50 | 157.50 | 157.85 |
| F | Q1, Q3 | 152.40, 162.60 | 149.90, 162.60 | 154.00, 162.60 | 154.28, 164.12 | 154.00, 162.60 |
| F | Min, Max | 135.9, 175.3 | 137.2, 174.0 | 135.9, 175.3 | 146.1, 170.2 | 135.9, 175.3 |
| F | Missing | 0 | 0 | 0 | 0 | 0 |
| M | n | 111 | 33 | 78 | 44 | 34 |
| M | Mean (SD) | 172.55 ( 7.946) | 173.03 ( 8.088) | 172.34 ( 7.929) | 172.91 ( 7.304) | 171.60 ( 8.729) |
| M | Median | 172.70 | 174.00 | 172.70 | 172.70 | 172.10 |
| M | Q1, Q3 | 168.25, 177.80 | 170.20, 177.80 | 167.60, 177.80 | 170.15, 177.80 | 165.42, 177.48 |
| M | Min, Max | 144.8, 195.6 | 144.8, 185.4 | 147.3, 195.6 | 147.3, 190.5 | 157.5, 195.6 |
| M | Missing | 0 | 0 | 0 | 0 | 0 |
For more information, it’s well worth your time to familiarize yourself with the select helpers that work with ‘dplyr’.
Sorting the Layers
Layers are one of the fundamental building blocks of
Tplyr. Each layer executes independently, and at the
end of a build they’re bound together. The ord_layer_index
variable allows you differentiate and sort layers after the table is
built. Layers are indexed in the order in which they were added to the
table using add_layer() or add_layers(). For
example, let’s say you wanted to reverse the order of the layers.
t %>%
select(starts_with("row"), starts_with("ord")) %>%
arrange(desc(ord_layer_index)) %>%
kable()| row_label1 | row_label2 | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|
| F | n | 2 | 1 | 1 |
| F | Mean (SD) | 2 | 1 | 2 |
| F | Median | 2 | 1 | 3 |
| F | Q1, Q3 | 2 | 1 | 4 |
| F | Min, Max | 2 | 1 | 5 |
| F | Missing | 2 | 1 | 6 |
| M | n | 2 | 2 | 1 |
| M | Mean (SD) | 2 | 2 | 2 |
| M | Median | 2 | 2 | 3 |
| M | Q1, Q3 | 2 | 2 | 4 |
| M | Min, Max | 2 | 2 | 5 |
| M | Missing | 2 | 2 | 6 |
| F | COMPLETED | 1 | 1 | 1 |
| F | DISCONTINUED | 1 | 1 | 2 |
| M | COMPLETED | 1 | 2 | 1 |
| M | DISCONTINUED | 1 | 2 | 2 |
Sorting the by Variables
Each by variable gets its own order column as well.
These will be named ord_layer_<n> where
<n> typically relates back to the
row_label variable (this isn’t necessarily the case when
count layers are nested - see vignette("count")).
These order variables will calculate based on the first applicable method below.
- If the
byvariable is a factor, the values of the ordering column will be associated with the factor levels. - If the variable has a
VARNvariable in thetargetdataset, (i.e.AVISIThasAVISITN, orPARAMhasPARAMN), that variable will be extracted and used as the ordering variable associated with that row label. - If neither 1 or 2 are true, the values in the ordering column will be based on an alphabetical sorting. The resulting column will be numeric.
Factor
If there’s no VARN variable in the target dataset,
Tplyr will then check if the variable you provided is a
factor. If you’re new to R, spending some time trying to understand
factor variables is quite worthwhile. Let’s look at example using the
variable ETHNIC and see some of the advantages in
practice.
tplyr_adsl$ETHNIC <- factor(tplyr_adsl$ETHNIC, levels=c("HISPANIC OR LATINO", "NOT HISPANIC OR LATINO", "DUMMMY"))
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_count(EOSSTT, by = ETHNIC)
) %>%
build() %>%
select(row_label1, row_label2, ord_layer_1) %>%
kable()| row_label1 | row_label2 | ord_layer_1 |
|---|---|---|
| HISPANIC OR LATINO | COMPLETED | 1 |
| HISPANIC OR LATINO | DISCONTINUED | 1 |
| NOT HISPANIC OR LATINO | COMPLETED | 2 |
| NOT HISPANIC OR LATINO | DISCONTINUED | 2 |
| DUMMMY | COMPLETED | 3 |
| DUMMMY | DISCONTINUED | 3 |
Factor variables have ‘levels’. These levels are essentially what the
VARN variables are trying to achieve - they specify the
order of the different values within the associated variable. The
variable we set above specifies that “HISPANIC OR LATINO” should sort
first, then “NOT HISPANIC OR LATINO”, and finally “DUMMY”. Notice how
they’re not alphabetical?
A highly advantageous aspect of using factor variables in Tplyr is that factor variables can be used to insert dummy values into your table. Consider this line of code from above:
tplyr_adsl$ETHNIC <- factor(tplyr_adsl$ETHNIC, levels=c("HISPANIC OR LATINO", "NOT HISPANIC OR LATINO", "DUMMMY"))This is converting the variable ETHNIC to a factor, then
setting the factor levels. But it doesn’t change any of the
values in the dataset - there are no values of “dummy” within
ETHNIC in ADSL. Yet in the output built above, you see rows
for “DUMMY”. By using factors, you can insert rows into your
Tplyr table that don’t exist in the data. This is
particularly helpful if you’re working with data early on in a study,
where certain values are expected, yet do not currently exist in the
data. This will help you prepare tables that are complete even when your
data are not.
VARN
To demonstrate the use of VARN sorting, consider the
variable RACE. In ADSL, RACE also
has RACEN:
| RACEN | RACE |
|---|---|
| 1 | WHITE |
| 2 | BLACK OR AFRICAN AMERICAN |
| 6 | AMERICAN INDIAN OR ALASKA NATIVE |
Tplyr will automatically figure this out for you,
and pull the RACEN values into the variable
ord_layer_1.
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_count(EOSSTT, by = RACE)
) %>%
build() %>%
select(row_label1, row_label2, ord_layer_1) %>%
arrange(ord_layer_1) %>%
kable()| row_label1 | row_label2 | ord_layer_1 |
|---|---|---|
| WHITE | COMPLETED | 1 |
| WHITE | DISCONTINUED | 1 |
| BLACK OR AFRICAN AMERICAN | COMPLETED | 2 |
| BLACK OR AFRICAN AMERICAN | DISCONTINUED | 2 |
| AMERICAN INDIAN OR ALASKA NATIVE | COMPLETED | 6 |
| AMERICAN INDIAN OR ALASKA NATIVE | DISCONTINUED | 6 |
Sorting Descriptive Statistic Summaries
After the by variables, each layer will sort results
slightly differently. We’ll start with the most simple case -
descriptive statistic layers. As the user, you have full control over
the order in which results present using
set_format_strings(). Results will be ordered based on the
order in which you create your f_str() objects.
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_desc(HEIGHTBL) %>%
set_format_strings(
'Group 1' = f_str('xx.x', mean),
'Group 2' = f_str('xx.x', median),
'Group 3' = f_str('xx.x', sd)
)
) %>%
build() %>%
select(starts_with("row"), starts_with("ord")) %>%
kable()| row_label1 | ord_layer_index | ord_layer_1 |
|---|---|---|
| Group 1 | 1 | 1 |
| Group 2 | 1 | 2 |
| Group 3 | 1 | 3 |
Each of the separate “Groups” added above were indexed based on their
position in set_format_strings(). If you’d like to change
the order, all you need to do is update your
set_format_strings() call.
Sorting Count Layers
The order in which results appear on a frequency table can be
deceptively complex and depends on the situation at hand. With this in
mind, Tplyr has 3 different methods of ordering the
results of a count layer using the function
set_order_count_method():
- “byfactor” - The default method is to sort by a factor. If the input variable is not a factor, alphabetical sorting will be used.
- “byvarn” - Similar to a ‘by’ variable, a count target can be sorted with a VARN variable existing in the target dataset.
-
“bycount” - This is the most complex method. Many
tables require counts to be sorted based on the counts within a
particular group, like a treatment variable. Tplyr can
populate the ordering column based on numeric values within any results
column. This requires some more granular control, for which we’ve
created the functions
set_ordering_cols()andset_result_order_var()to specify the column and numeric value on which the ordering column should be based.
“byfactor” and “byvarn”
“byfactor” is the default ordering method of results for count
layers. Both “byfactor” and “byvarn” behave exactly like the order
variables associated with by variables in a
Tplyr table. For “byvarn”, you must set the sort method
using set_order_count_method().
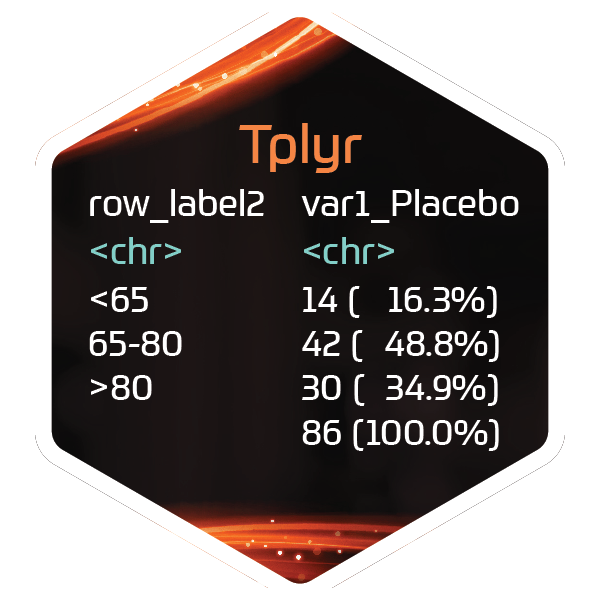
tplyr_adsl$AGEGR1 <- factor(tplyr_adsl$AGEGR1, c("<65", "65-80", ">80"))
# Warnings suppressed to remove 'forcats' implicit NA warning
suppressWarnings({
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_count(AGEGR1) %>%
# This is the default and not needed
set_order_count_method("byfactor")
) %>%
build() %>%
select(row_label1, ord_layer_1) %>%
kable()
})| row_label1 | ord_layer_1 |
|---|---|
| <65 | 1 |
| 65-80 | 2 |
| >80 | 3 |
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_count(RACE) %>%
set_order_count_method("byvarn")
) %>%
build() %>%
select(row_label1, ord_layer_1) %>%
kable()| row_label1 | ord_layer_1 |
|---|---|
| AMERICAN INDIAN OR ALASKA NATIVE | 6 |
| BLACK OR AFRICAN AMERICAN | 2 |
| WHITE | 1 |
“bycount”
Using count-based sorting is where things get more complicated. There are multiple items to consider:
- What column do you want to sort by?
- If there are multiple numbers in the column, like “n (%) [event]” type tables, which number should be used to create the sort variable?
We’ve created helper functions to aid in making this step more
intuitive from a user perspective, and to maintain the flexibility that
you need. The two functions that you need here are
set_ordering_cols() and
set_result_order_var().
tplyr_table(tplyr_adae, TRTA) %>%
add_layer(
group_count(AEDECOD) %>%
# This will present 3 numbers in a cell
set_format_strings(f_str("xx (xx.x%) [x]", distinct_n, distinct_pct, n)) %>%
# This makes the distinct numbers available
set_distinct_by(USUBJID) %>%
# Choosing "bycount" ordering for the result variable
set_order_count_method("bycount") %>%
# This will target the results column for Xanomeline High Dose, or `var1_Xanomeline High Dose`
set_ordering_cols("Xanomeline High Dose") %>%
# The number we want to pull out is the distinct N counts
set_result_order_var(distinct_n)
) %>%
build() %>%
arrange(desc(ord_layer_1)) %>%
select(row_label1, `var1_Xanomeline High Dose`, ord_layer_1) %>%
head() %>%
kable()| row_label1 | var1_Xanomeline High Dose | ord_layer_1 |
|---|---|---|
| PRURITUS | 26 (61.9%) [38] | 26 |
| ERYTHEMA | 14 (33.3%) [22] | 14 |
| RASH | 11 (26.2%) [18] | 11 |
| HYPERHIDROSIS | 8 (19.0%) [10] | 8 |
| SKIN IRRITATION | 5 (11.9%) [8] | 5 |
| RASH PRURITIC | 2 ( 4.8%) [3] | 2 |
In the above example, the results columns of the output table
actually contain three different numbers: the distinct counts, the
distinct percentage, and the non-distinct counts. We want to use
distinct counts, so we choose distinct_n.
The next question that we need to answer when sorting by counts is which result column to take counts out of. Here, we have three results columns - one for each treatment group in the dataset. We want to use the results for the treatment group “Xanomeline High Dose”, so we provide the name of the treatment group.
But what if you have an additional column variable on top of the treatment groups?
tplyr_table(tplyr_adae, TRTA, cols=SEX) %>%
add_layer(
group_count(AEDECOD) %>%
# This will present 3 numbers in a cell
set_format_strings(f_str("xx (xx.x%) [x]", distinct_n, distinct_pct, n)) %>%
# This makes the distinct numbers available
set_distinct_by(USUBJID) %>%
# Choosing "bycount" ordering for the result variable
set_order_count_method("bycount") %>%
# This will target the results column for Xanomeline High Dose, or `var1_Xanomeline High Dose`
set_ordering_cols("Xanomeline High Dose", "F") %>%
# The number we want to pull out is the distinct N counts
set_result_order_var(distinct_n)
) %>%
build() %>%
arrange(desc(ord_layer_1)) %>%
select(row_label1, `var1_Xanomeline High Dose_F`, ord_layer_1) %>%
head() %>%
kable()| row_label1 | var1_Xanomeline High Dose_F | ord_layer_1 |
|---|---|---|
| PRURITUS | 11 (78.6%) [14] | 11 |
| ERYTHEMA | 7 (50.0%) [8] | 7 |
| RASH | 3 (21.4%) [5] | 3 |
| HYPERHIDROSIS | 2 (14.3%) [2] | 2 |
| RASH PRURITIC | 1 ( 7.1%) [1] | 1 |
| SKIN IRRITATION | 1 ( 7.1%) [2] | 1 |
Here we’re ordering on the female subjects in the “Xanomeline High
Dose” cohort. In set_result_order_var(), you need to enter
the values from each variable between treat_var and any
variable entered in cols that you’d like to extract.
Nested Sorting
Nested count layers add one more piece to the puzzle. As a reminder, nested count layers are count summaries that are summarizing both a grouping variable, and a variable that’s being grouped. The best example is probably Adverse Event tables, where we want to see adverse events that occurred within different body systems.
tplyr_table(tplyr_adae, TRTA) %>%
add_layer(
group_count(vars(AEBODSYS, AEDECOD))
) %>%
build() %>%
head() %>%
kable()| row_label1 | row_label2 | var1_Placebo | var1_Xanomeline High Dose | var1_Xanomeline Low Dose | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|---|---|---|
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | SKIN AND SUBCUTANEOUS TISSUE DISORDERS | 47 (100.0%) | 111 (100.0%) | 118 (100.0%) | 1 | 1 | Inf |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | ACTINIC KERATOSIS | 0 ( 0.0%) | 1 ( 0.9%) | 0 ( 0.0%) | 1 | 1 | 1 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | ALOPECIA | 1 ( 2.1%) | 0 ( 0.0%) | 0 ( 0.0%) | 1 | 1 | 2 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | BLISTER | 0 ( 0.0%) | 2 ( 1.8%) | 8 ( 6.8%) | 1 | 1 | 3 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | COLD SWEAT | 3 ( 6.4%) | 0 ( 0.0%) | 0 ( 0.0%) | 1 | 1 | 4 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | DERMATITIS ATOPIC | 1 ( 2.1%) | 0 ( 0.0%) | 0 ( 0.0%) | 1 | 1 | 5 |
In a layer that uses nesting, we need one more order variable - as we’re now concerned with the sorting of both the outside and inside variable. Counts are being summarized for both - so we need to know how both should be sorted. Additionally, we need to make sure that, in this case, the adverse events within a body system stay within the rows of that body system.
These result variables will always be the last two order variables
output by Tplyr. In the above example,
ord_layer_1 is for AEBODSYS and
ord_layer_2 is for AEDECOD. Note that
ord_layer_2 has Inf where
row_label1 and row_label2 are both equal. This
is the row that summarizes the AEBODSYS counts. By default,
Tplyr is set to assume that you will use
descending sort on the order variable associated with
the inside count variable (i.e. AEDECOD). This is because
in nested count layer you will often want to sort by descending
occurrence of the inside target variable. If you’d like to use ascending
sorting instead, we offer the function
set_outer_sort_position().
tplyr_table(tplyr_adae, TRTA) %>%
add_layer(
group_count(vars(AEBODSYS, AEDECOD)) %>%
set_outer_sort_position("asc")
) %>%
build() %>%
arrange(ord_layer_1, ord_layer_2) %>%
select(starts_with("row"), starts_with("ord_layer")) %>%
head() %>%
kable()| row_label1 | row_label2 | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | SKIN AND SUBCUTANEOUS TISSUE DISORDERS | 1 | 1 | -Inf |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | ACTINIC KERATOSIS | 1 | 1 | 1 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | ALOPECIA | 1 | 1 | 2 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | BLISTER | 1 | 1 | 3 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | COLD SWEAT | 1 | 1 | 4 |
| SKIN AND SUBCUTANEOUS TISSUE DISORDERS | DERMATITIS ATOPIC | 1 | 1 | 5 |
Notice that the Inf has now switched to
-Inf to ensure that the AEBODSYS row stays at
the top of the group.
Another consideration of nested sorting is whether or not you want to
sort both result variables the same way. Do you want to sort both by
counts? Or do you want to sort one alphabetically and the other by
count? Or maybe one has a VARN variable associated with it?
For this reason, set_order_count_method() can take in a
2-element character vector, where the first element specifies the
outside variable and the second the inside variable.
tplyr_table(tplyr_adsl, TRT01A) %>%
add_layer(
group_count(vars(EOSSTT, DCDECOD)) %>%
set_order_count_method(c("byfactor", "bycount"))
) %>%
build() %>%
select(starts_with("row"), starts_with("ord")) %>%
kable()| row_label1 | row_label2 | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|
| COMPLETED | COMPLETED | 1 | 1 | Inf |
| COMPLETED | COMPLETED | 1 | 1 | 58 |
| DISCONTINUED | DISCONTINUED | 1 | 2 | Inf |
| DISCONTINUED | ADVERSE EVENT | 1 | 2 | 8 |
| DISCONTINUED | DEATH | 1 | 2 | 2 |
| DISCONTINUED | LACK OF EFFICACY | 1 | 2 | 3 |
| DISCONTINUED | LOST TO FOLLOW-UP | 1 | 2 | 1 |
| DISCONTINUED | PHYSICIAN DECISION | 1 | 2 | 1 |
| DISCONTINUED | PROTOCOL VIOLATION | 1 | 2 | 2 |
| DISCONTINUED | STUDY TERMINATED BY SPONSOR | 1 | 2 | 2 |
| DISCONTINUED | WITHDRAWAL BY SUBJECT | 1 | 2 | 9 |
In the example above, EOSTT is ordered alphabetically
(recall that using “byfactor” when the variable is not a factor will do
alphabetical sorting), and DSDECOD is ordered by count.
If only one method is provided, that method will automatically be
applied to both variables. So in the example below, “bycount” is applied
to both EOSTT and DSDECOD.
tplyr_table(tplyr_adsl, TRT01A) %>%
add_total_group() %>%
add_layer(
group_count(vars(EOSSTT, DCDECOD)) %>%
set_order_count_method("bycount") %>%
#set_order_count_method("bycount", "bycount") %>% This is functionally the same.
set_ordering_cols(Total)
) %>%
build() %>%
select(starts_with("row"), var1_Total, starts_with("ord")) %>%
kable()| row_label1 | row_label2 | var1_Total | ord_layer_index | ord_layer_1 | ord_layer_2 |
|---|---|---|---|---|---|
| COMPLETED | COMPLETED | 110 ( 43.3%) | 1 | 110 | Inf |
| COMPLETED | COMPLETED | 110 ( 43.3%) | 1 | 110 | 110 |
| DISCONTINUED | DISCONTINUED | 144 ( 56.7%) | 1 | 144 | Inf |
| DISCONTINUED | ADVERSE EVENT | 92 ( 36.2%) | 1 | 144 | 92 |
| DISCONTINUED | DEATH | 3 ( 1.2%) | 1 | 144 | 3 |
| DISCONTINUED | LACK OF EFFICACY | 4 ( 1.6%) | 1 | 144 | 4 |
| DISCONTINUED | LOST TO FOLLOW-UP | 2 ( 0.8%) | 1 | 144 | 2 |
| DISCONTINUED | PHYSICIAN DECISION | 3 ( 1.2%) | 1 | 144 | 3 |
| DISCONTINUED | PROTOCOL VIOLATION | 6 ( 2.4%) | 1 | 144 | 6 |
| DISCONTINUED | STUDY TERMINATED BY SPONSOR | 7 ( 2.8%) | 1 | 144 | 7 |
| DISCONTINUED | WITHDRAWAL BY SUBJECT | 27 ( 10.6%) | 1 | 144 | 27 |
Sorting Shift Tables
Shift tables keep things relatively simple when it comes to sorting and use the “byfactor” method seen above. We encourage this primarily because you likely want the benefits of factor variables on a shift layer. For example, consider this table:
tplyr_table(tplyr_adlb, TRTA, where=PARAMCD == "CK") %>%
add_layer(
group_shift(vars(row = BNRIND, column = ANRIND), by = vars(PARAM, AVISIT))
) %>%
build() %>%
select(-starts_with('var1')) %>%
head(20) %>%
kable()| row_label1 | row_label2 | row_label3 | ord_layer_index | ord_layer_1 | ord_layer_2 | ord_layer_3 |
|---|---|---|---|---|---|---|
| Creatine Kinase (U/L) | Week 12 | H | 1 | 35 | 12 | 1 |
| Creatine Kinase (U/L) | Week 12 | N | 1 | 35 | 12 | 3 |
| Creatine Kinase (U/L) | Week 24 | H | 1 | 35 | 24 | 1 |
| Creatine Kinase (U/L) | Week 24 | N | 1 | 35 | 24 | 3 |
| Creatine Kinase (U/L) | Week 8 | H | 1 | 35 | 8 | 1 |
| Creatine Kinase (U/L) | Week 8 | N | 1 | 35 | 8 | 3 |
There are a few problems here:
- “H” sorts before “N” alphabetically
- We’re missing the rows for “L” on most visits, even though “L” in in
the data for
BNRIND.
Using factor variables cleans this right up for us:
tplyr_adlb$BNRIND <- factor(tplyr_adlb$BNRIND, levels=c("L", "N", "H"))
tplyr_adlb$ANRIND <- factor(tplyr_adlb$ANRIND, levels=c("L", "N", "H"))
tplyr_table(tplyr_adlb, TRTA, where=PARAMCD == "CK") %>%
add_layer(
group_shift(vars(row = BNRIND, column = ANRIND), by = vars(PARAM, AVISIT))
) %>%
build() %>%
select(-starts_with('var1')) %>%
head(20) %>%
kable()| row_label1 | row_label2 | row_label3 | ord_layer_index | ord_layer_1 | ord_layer_2 | ord_layer_3 |
|---|---|---|---|---|---|---|
| Creatine Kinase (U/L) | Week 12 | L | 1 | 35 | 12 | 1 |
| Creatine Kinase (U/L) | Week 12 | N | 1 | 35 | 12 | 2 |
| Creatine Kinase (U/L) | Week 12 | H | 1 | 35 | 12 | 3 |
| Creatine Kinase (U/L) | Week 24 | L | 1 | 35 | 24 | 1 |
| Creatine Kinase (U/L) | Week 24 | N | 1 | 35 | 24 | 2 |
| Creatine Kinase (U/L) | Week 24 | H | 1 | 35 | 24 | 3 |
| Creatine Kinase (U/L) | Week 8 | L | 1 | 35 | 8 | 1 |
| Creatine Kinase (U/L) | Week 8 | N | 1 | 35 | 8 | 2 |
| Creatine Kinase (U/L) | Week 8 | H | 1 | 35 | 8 | 3 |
Now we have the nice “L”, “N”, “H” order that we’d like to see. Other sort methods on a shift table are fairly unlikely, as the matrix structure of the counts displayed by shift tables is relevant to the presentation and interpreting results.
Happy sorting!