Introduction
This vignette will explore in detail all the possibilities of the xportr package for applying information from a metadata object to an R created dataset using the core xportr functions.
We will also explore the following:
- What goes in a Submission to a Health Authority, and what role does xportr play in that Submission?
- What is xportr validating behind the scenes?
- Breakdown of xportr and a ADaM dataset specification file.
- Using
options()andxportr_metadata()to enhance your xportr experience. - Understanding the warning and error messages for each xportr function.
- A brief discussion on future work.
NOTE: We use the phrase metadata object throughout this package. A metadata object can either be a specification file read into R as a dataframe or a metacore object. The metadata object created via the metacore package has additional features not covered here, but at its core is using a specification file. However, xportr will work with either a dataframe or a metacore object.
What goes in a Submission to a Health Authority?
Quite a bit! We will focus on the data deliverables and supporting documentation needed for a successful submission to a Health Authority and how xportr can play a key role. We will briefly look at three parts:
- Study Data Standardization Plan
- SDTM Data Package
- ADaM Data Package
Study Data Standardization Plan
The Study Data Standardization Plan (SDSP) establishes and documents a plan for describing the data standardization approach for clinical and nonclinical studies within a development program. The SDSP also assists the FDA in identifying potential data standardization issues early in the development program. We hope the brevity of this section does not belie the huge importance of this document. Please see Study Data Standardisation Plan (SDSP) Package maintained by the PHUSE working group. However, we want to focus more on the actual data and how xportr can play a role in the submission.
SDTM and ADaM Data Packages
SDTM: The primary pieces of the SDTM package are the SDTM annotated case report forms (acrf.pdf), the data definitions document (define.xml), the Study Data Reviewer’s Guide (sdrg.pdf) and the datasets in xpt Version 5 format. The Version 5 xpt file is the required submission format for all datasets going to the Health Authorities.
ADaM: The key components of the ADaM package are very similar to SDTM package with a few additions: define.xml, Analysis Study Data Reviewer’s Guide (adrg.pdf), Analysis Results Metadata (analysis-results-metadata.pdf) and datasets as Version 5 xpt format.
As both Data Packages need compliant xpt files, we feel
that xportr can play a pivotal role here. The core
functions in xportr can be used to apply information from
the metadata object to the datasets giving users feedback on
the quality of the metadata and data. xportr_write() can
then be used to write out the final dataset as an xpt file,
which can be submitted to a Health Authority.
What is {xportr} validating in these Data
Packages?
The xpt Version 5 files form the backbone of any
successful Submission and are govern by quite a lot of rules and
suggested guidelines. As you are preparing your data packages for
submission the suite of core xportr functions, plus
xportr_write(), helps to check that your datasets are
submission compliant. The package checks many of the latest rules laid
out in the Study
Data Technical Conformance Guide, but please note that it is not yet
an exhaustive list of checks. We envision that users are also submitting
their xpts and metadata to additional validation
software.
Each of the core xportr functions for applying labels,
types, formats, order and lengths provides feedback to users on
submission compliance. However, a final check is implemented when
xportr_write() is called to create the xpt.
xportr_write() calls xpt_validate(),
which is a behind the scenes/non-exported function that does a final
check for compliance. At the time of {xportr} v0.3.0 we are
checking the following when a user writes out an xpt
file.:
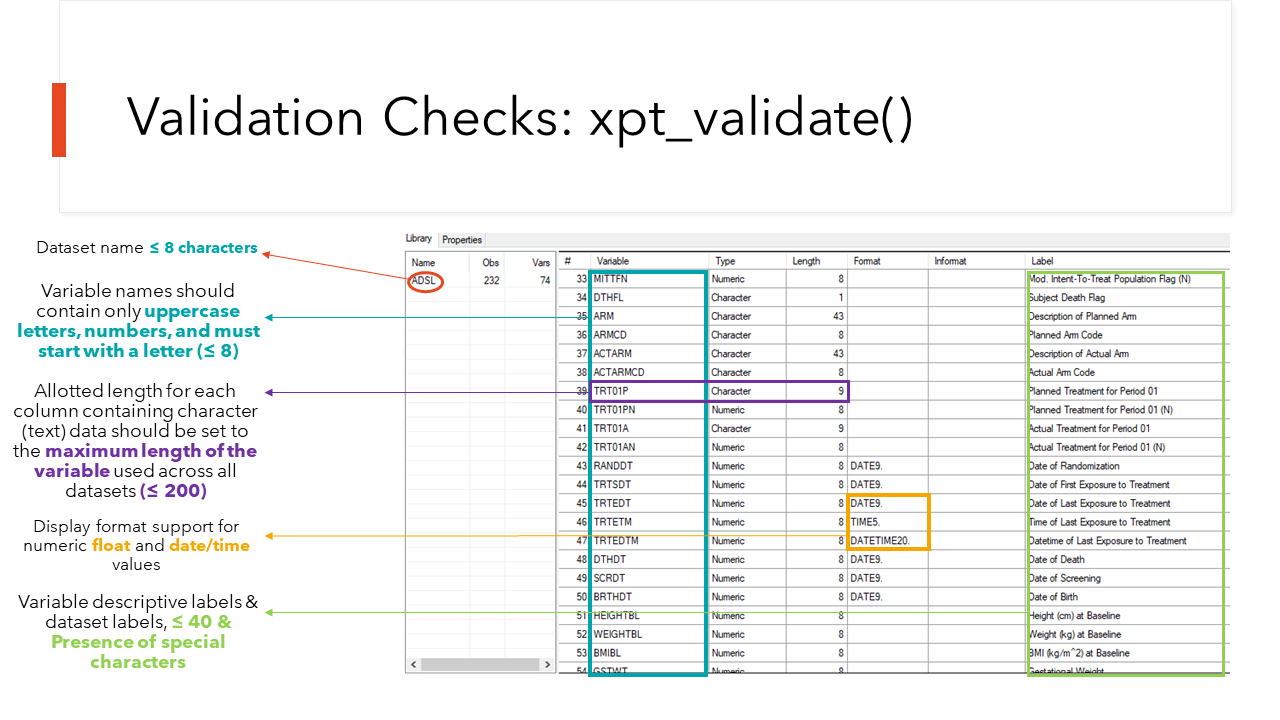
{xportr} in action
In this section, we are going to explore the 5 core xportr functions using:
-
data("adsl_xportr", package = "xportr")- An ADSL ADaM dataset from the Pilot 3 Submission to the FDA -
data("var_spec", package = "xportr")- The ADSL ADaM Specification File from the Pilot 3 Submission to the FDA
We will focus on warning and error messaging with contrived examples from these functions by manipulating either the datasets or the specification files.
NOTE: We have made the ADSL and Spec available in
this package. Users can find additional datasets and specification files
on our repo in
the example_data_specs folder. This is to keep the package
to a minimum size.
Using options() and xportr_metadata() to
enhance your experience.
Before we dive into the functions, we want to point out some quality
of life utilities to make your xpt generation life a little
bit easier.
NOTE: As long as you have a well-defined
metadata object you do NOT need to use options()
or xportr_metadata(), but we find these handy to use and
think they deserve a quick mention!
You’ve got options() or
xportr_options()
xportr is built with certain assumptions around
specification column names and information in those columns. We have
found that each company specification file can differ slightly from our
assumptions. For example, one company might call a column
Variables, another Variable and another
variables. Rather than trying to regex ourselves out of
this situation, we have introduced options().
options() allows users to control those assumptions inside
xportr functions based on their needs.
Additionally, we have a helper function xportr_options()
which works just like the options() but, it can also be
used to get the current state of the xportr options.
Let’s take a look at our example specification file names available
in this package. We can see that all the columns start with an upper
case letter and have spaces in several of them. We could convert all the
column names to lower case and deal with the spacing using some
dplyr functions or base R, or we could just use
options()!
library(xportr)
library(dplyr)
library(haven)
data("adsl_xportr", "var_spec", "dataset_spec", package = "xportr")
colnames(var_spec)
[1] "Order" "Dataset" "Variable"
[4] "Label" "Data Type" "Length"
[7] "Significant Digits" "Format" "Mandatory"
[10] "Assigned Value" "Codelist" "Common"
[13] "Origin" "Pages" "Method"
[16] "Predecessor" "Role" "Comment"
[19] "Developer Notes"
ADSL <- adsl_xportrBy using options() or xportr_options() at
the beginning of our script we can tell xportr what the
valid names are (see chunk below). Please note that before we set the
options the package assumed every thing was in lowercase and there were
no spaces in the names. After running options() or
xportr_options(), xportr sees the column
Variable as the valid name rather than
variable. You can inspect xportr_options
function docs to look at additional options.
xportr_options(
xportr.variable_name = "Variable",
xportr.label = "Label",
xportr.type_name = "Data Type",
xportr.format = "Format",
xportr.length = "Length",
xportr.order_name = "Order"
)
# Or alternatively
options(
xportr.variable_name = "Variable",
xportr.label = "Label",
xportr.type_name = "Data Type",
xportr.format = "Format",
xportr.length = "Length",
xportr.order_name = "Order"
)Are we being too verbose?
One final note on the options. 4 of the core xportr
functions have the ability to set messaging as
"none", "message", "warn", "stop". Setting each of these in
all your calls can be a bit repetitive. You can use
options() or xportr_options() to set these at
a higher level and avoid this repetition.
# Default verbose is set to `none`
xportr_options(
xportr.format_verbose = "none",
xportr.label_verbose = "none",
xportr.length_verbose = "none",
xportr.type_verbose = "none"
)
xportr_options(
xportr.format_verbose = "none", # Disables any messaging, keeping the console output clean
xportr.label_verbose = "message", # Sends a standard message to the console
xportr.length_verbose = "warn", # Sends a warning message to the console
xportr.type_verbose = "stop" # Stops execution and sends an error message to the console
)Going meta
Each of the core xportr functions requires several inputs: A valid dataframe, a metadata object and a domain name, along with optional messaging. For example, here is a simple call using all of the functions. As you can see, a lot of information is repeated in each call.
ADSL %>%
xportr_type(var_spec, "ADSL", "message") %>%
xportr_length(var_spec, "ADSL", verbose = "message") %>%
xportr_label(var_spec, "ADSL", "message") %>%
xportr_order(var_spec, "ADSL", "message") %>%
xportr_format(var_spec, "ADSL") %>%
xportr_df_label(dataset_spec, "ADSL") %>%
xportr_write("adsl.xpt")To help reduce these repetitive calls, we have created
xportr_metadata(). A user can just set the
metadata object and the Domain name in the first call, and this
will be passed on to the other functions. Much cleaner!
ADSL %>%
xportr_metadata(var_spec, "ADSL") %>%
xportr_type() %>%
xportr_length(length_source = "metadata") %>%
xportr_label() %>%
xportr_order() %>%
xportr_format() %>%
xportr_df_label(dataset_spec) %>%
xportr_write("adsl.xpt")Warnings and Errors
For the next six sections, we are going to explore the Warnings and Errors messages generated by the xportr core functions. To better explore these, we will either manipulate the ADaM dataset or specification file to help showcase the ability of the xportr functions to detect issues.
NOTE: We have made the ADSL,
xportr::adsl, and Specification File,
xportr::var_spec, available in this package. Users can find
additional datasets and specification files on our repo in the
example_data_specs folder.
Setting up our metadata object
First, let’s read in the specification file and call it
var_spec. Note that we are not using options()
here. We will do some slight manipulation to the column names by doing
all lower case, and changing Data Type to type
and making the Order column numeric. You can also use
options() for this step as well. The var_spec
object has five dataset specification files stacked on top of each
other. We will make use of the ADSL subset of
var_spec. You can make use of the Search field above the
dataset column to subset the specification file for ADSL
Similarly, we can read the Dataset spec file and call it
dataset_spec.
var_spec <- var_spec %>%
rename(type = "Data Type") %>%
rename_with(tolower)
dataset_spec <- dataset_spec %>%
rename(label = "Description") %>%
rename_with(tolower)
xportr_type()
We are going to explore the type column in the metadata object. A
submission to a Health Authority should only have character and numeric
types in the data. In the ADSL data we have several columns
that are in the Date type: TRTSDT, TRTEDT,
SCRFDT, EOSDT, FRVDT,
RANDDT, DTHDT, LSTALVDT - under
the hood these are actually numeric values and will be left as is. We
will change one variable type to a factor variable, which is a
common data structure in R, to give us some educational opportunities to
see xportr_type() in action.
Rows: 306
Columns: 9
$ STUDYID <fct> CDISCPILOT01, CDISCPILOT01, CDISCPILOT01, CDISCPILOT01, CDISC…
$ TRTSDT <date> 2014-01-02, 2012-08-05, 2013-07-19, 2014-03-18, 2014-07-01, …
$ TRTEDT <date> 2014-07-02, 2012-09-01, 2014-01-14, 2014-03-31, 2014-12-30, …
$ SCRFDT <date> NA, NA, NA, NA, NA, NA, 2013-12-20, NA, NA, NA, NA, NA, NA, …
$ EOSDT <date> 2014-07-02, 2012-09-02, 2014-01-14, 2014-04-14, 2014-12-30, …
$ FRVDT <date> NA, 2013-02-18, NA, 2014-09-15, NA, 2013-07-28, NA, NA, 2013…
$ RANDDT <date> 2014-01-02, 2012-08-05, 2013-07-19, 2014-03-18, 2014-07-01, …
$ DTHDT <date> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ LSTALVDT <date> 2014-07-02, 2012-09-02, 2014-01-14, 2014-04-14, 2014-12-30, …adsl_type <- xportr_type(.df = adsl_fct, metadata = var_spec, domain = "ADSL", verbose = "warn")
── Variable type mismatches found. ──
✔ 1 variable coerced
Warning: Variable type(s) in dataframe don't match metadata: `STUDYID`
- `STUDYID` was coerced to <character>. (type in data: factor, type in metadata: text)
i Types in metadata considered as character (xportr.character_metadata_types option): 'character', 'char', 'text', 'date', 'posixct', 'posixt', 'datetime', 'time', 'partialdate', 'partialtime', 'partialdatetime', 'incompletedatetime', 'durationdatetime', and 'intervaldatetime'
i Types in metadata considered as numeric (xportr.numeric_metadata_types option): 'integer', 'numeric', 'num', and 'float'
i Types in data considered as character (xportr.character_types option): 'character'
i Types in data considered as numeric (xportr.numeric_types option): 'integer', 'float', 'numeric', 'posixct', 'posixt', 'time', 'date', and 'hms'Success! As we can see below, xportr_type() applied the
types from the metadata object to the STUDYID variables
converting to the proper type. The functions in xportr
also display this coercion to the user in the console, which is seen
above.
glimpse(adsl_type_glimpse)
Rows: 306
Columns: 9
$ STUDYID <chr> "CDISCPILOT01", "CDISCPILOT01", "CDISCPILOT01", "CDISCPILOT01…
$ TRTSDT <date> 2014-01-02, 2012-08-05, 2013-07-19, 2014-03-18, 2014-07-01, …
$ TRTEDT <date> 2014-07-02, 2012-09-01, 2014-01-14, 2014-03-31, 2014-12-30, …
$ SCRFDT <date> NA, NA, NA, NA, NA, NA, 2013-12-20, NA, NA, NA, NA, NA, NA, …
$ EOSDT <date> 2014-07-02, 2012-09-02, 2014-01-14, 2014-04-14, 2014-12-30, …
$ FRVDT <date> NA, 2013-02-18, NA, 2014-09-15, NA, 2013-07-28, NA, NA, 2013…
$ RANDDT <date> 2014-01-02, 2012-08-05, 2013-07-19, 2014-03-18, 2014-07-01, …
$ DTHDT <date> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ LSTALVDT <date> 2014-07-02, 2012-09-02, 2014-01-14, 2014-04-14, 2014-12-30, …Note that xportr_type(verbose = "warn") was set so the
function has provided feedback, which would show up in the console, on
which variables were converted as a warning message. However, you can
set verbose = "stop" so that the types are not applied if
the data does not match what is in the specification file. Using
verbose = "stop" will instantly stop the processing of this
function and not create the object. A user will need to alter the
variables in their R script before using xportr_type()
adsl_type <- xportr_type(.df = adsl_fct, metadata = var_spec, domain = "ADSL", verbose = "stop")
── Variable type mismatches found. ──
✔ 1 variable coerced
Error in `xportr_logger()` at xportr/R/messages.R:126:5:
! Variable type(s) in dataframe don't match metadata: `STUDYID`
- `STUDYID` was coerced to <character>. (type in data: factor, type in metadata: text)
i Types in metadata considered as character (xportr.character_metadata_types option): 'character', 'char', 'text', 'date', 'posixct', 'posixt', 'datetime', 'time', 'partialdate', 'partialtime', 'partialdatetime', 'incompletedatetime', 'durationdatetime', and 'intervaldatetime'
i Types in metadata considered as numeric (xportr.numeric_metadata_types option): 'integer', 'numeric', 'num', and 'float'
i Types in data considered as character (xportr.character_types option): 'character'
i Types in data considered as numeric (xportr.numeric_types option): 'integer', 'float', 'numeric', 'posixct', 'posixt', 'time', 'date', and 'hms'
xportr_length()
There are two sources of length (data-driven and spec-driven):
Data-driven length: max length for character columns and 8 for other data types
Spec-driven length: from the metadata
- Users can either specify the length in the metadata or leave it blank for data-driven length.
- When the length is missing in the metadata, the data-driven length will be applied.
Next we will use xportr_length() to apply the length
column of the metadata object to the ADSL dataset.
Using the str() function we have displayed all the
variables with their attributes. You can see that each variable has a
label, but there is no information on the lengths of the variable.
tibble [306 × 51] (S3: tbl_df/tbl/data.frame)
$ STUDYID : chr [1:306] "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" ...
..- attr(*, "label")= chr "Study Identifier"
$ USUBJID : chr [1:306] "01-701-1015" "01-701-1023" "01-701-1028" "01-701-1033" ...
..- attr(*, "label")= chr "Unique Subject Identifier"
$ SUBJID : chr [1:306] "1015" "1023" "1028" "1033" ...
..- attr(*, "label")= chr "Subject Identifier for the Study"
$ RFSTDTC : chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Subject Reference Start Date/Time"
$ RFENDTC : chr [1:306] "2014-07-02" "2012-09-02" "2014-01-14" "2014-04-14" ...
..- attr(*, "label")= chr "Subject Reference End Date/Time"
$ RFXSTDTC: chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Date/Time of First Study Treatment"
$ RFXENDTC: chr [1:306] "2014-07-02" "2012-09-01" "2014-01-14" "2014-03-31" ...
..- attr(*, "label")= chr "Date/Time of Last Study Treatment"
$ RFICDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Informed Consent"
$ RFPENDTC: chr [1:306] "2014-07-02T11:45" "2013-02-18" "2014-01-14T11:10" "2014-09-15" ...
..- attr(*, "label")= chr "Date/Time of End of Participation"
$ DTHDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Death"
$ DTHFL : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Subject Death Flag"
$ SITEID : chr [1:306] "701" "701" "701" "701" ...
..- attr(*, "label")= chr "Study Site Identifier"
$ AGE : num [1:306] 63 64 71 74 77 85 59 68 81 84 ...
..- attr(*, "label")= chr "Age"
$ AGEU : chr [1:306] "YEARS" "YEARS" "YEARS" "YEARS" ...
..- attr(*, "label")= chr "Age Units"
$ SEX : chr [1:306] "F" "M" "M" "M" ...
..- attr(*, "label")= chr "Sex"
$ RACE : chr [1:306] "WHITE" "WHITE" "WHITE" "WHITE" ...
..- attr(*, "label")= chr "Race"
$ ETHNIC : chr [1:306] "HISPANIC OR LATINO" "HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" ...
..- attr(*, "label")= chr "Ethnicity"
$ ARMCD : chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Planned Arm Code"
$ ARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
$ ACTARMCD: chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Actual Arm Code"
$ ACTARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
$ COUNTRY : chr [1:306] "USA" "USA" "USA" "USA" ...
..- attr(*, "label")= chr "Country"
$ DMDTC : chr [1:306] "2013-12-26" "2012-07-22" "2013-07-11" "2014-03-10" ...
..- attr(*, "label")= chr "Date/Time of Collection"
$ DMDY : num [1:306] -7 -14 -8 -8 -7 -21 NA -9 -13 -7 ...
..- attr(*, "label")= chr "Study Day of Collection"
$ TRT01P : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
$ TRT01A : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
$ TRTSDTM : POSIXct[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTSTMF : chr [1:306] "H" "H" "H" "H" ...
$ TRTEDTM : POSIXct[1:306], format: "2014-07-02 23:59:59" "2012-09-01 23:59:59" ...
$ TRTETMF : chr [1:306] "H" "H" "H" "H" ...
$ TRTSDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTEDT : Date[1:306], format: "2014-07-02" "2012-09-01" ...
$ TRTDURD : num [1:306] 182 28 180 14 183 26 NA 190 10 55 ...
$ SCRFDT : Date[1:306], format: NA NA ...
$ EOSDT : Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ EOSSTT : chr [1:306] "COMPLETED" "DISCONTINUED" "COMPLETED" "DISCONTINUED" ...
$ FRVDT : Date[1:306], format: NA "2013-02-18" ...
$ RANDDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ DTHDT : Date[1:306], format: NA NA ...
$ DTHDTF : chr [1:306] NA NA NA NA ...
$ DTHADY : num [1:306] NA NA NA NA NA NA NA NA NA NA ...
$ LDDTHELD: num [1:306] NA NA NA NA NA NA NA NA NA NA ...
$ LSTALVDT: Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ SAFFL : chr [1:306] "Y" "Y" "Y" "Y" ...
$ RACEGR1 : chr [1:306] "White" "White" "White" "White" ...
$ AGEGR1 : chr [1:306] "18-64" "18-64" ">64" ">64" ...
$ REGION1 : chr [1:306] "NA" "NA" "NA" "NA" ...
$ LDDTHGR1: chr [1:306] NA NA NA NA ...
$ DTH30FL : chr [1:306] NA NA NA NA ...
$ DTHA30FL: chr [1:306] NA NA NA NA ...
$ DTHB30FL: chr [1:306] NA NA NA NA ...
- attr(*, "label")= chr "Demographics"adsl_length <- xportr_length(
.df = ADSL,
metadata = var_spec,
domain = "ADSL",
verbose = "warn",
length_source = "metadata"
)
── Variable lengths missing from metadata. ──
✔ 30 lengths resolved `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
Warning: Variable(s) present in dataframe but doesn't exist in
`metadata`.Problem with `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`,
`DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`,
`TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`,
`DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`,
`DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
Warning: Variable(s) present in `metadata` but don't exist in dataframe.
✖ Problem with `SITEGR1`, `TRT01PN`, `TRT01AN`, `AVGDD`, `CUMDOSE`, `AGEGR1N`, `RACEN`, `ITTFL`, `EFFFL`, `COMP8FL`, `COMP16FL`, `COMP24FL`, `DISCONFL`, `DSRAEFL`, `BMIBL`, `BMIBLGR1`, `HEIGHTBL`, `WEIGHTBL`, `EDUCLVL`, `DISONSDT`, `DURDIS`, `DURDSGR1`, `VISIT1DT`, `VISNUMEN`, `RFENDT`, `DCDECOD`, `DCSREAS`, and `MMSETOT`Using xportr_length() with verbose = "warn"
we can apply the length column to all the columns in the dataset. The
function detects 30 variables, including RFXSTDTC and
DTHDTC, that are missing length from the metadata file.
However, lengths are still applied. For example, RFXSTDTC
and DTHDTC are given a length of 10. It also detects 28
variables that are present in the metadata but not in the dataset.
Using the str() function, you can see below that
xportr_length() successfully applied all the lengths of the
variable to the variables in the dataset.
tibble [306 × 51] (S3: tbl_df/tbl/data.frame)
$ STUDYID : chr [1:306] "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" ...
..- attr(*, "label")= chr "Study Identifier"
..- attr(*, "width")= num 12
$ USUBJID : chr [1:306] "01-701-1015" "01-701-1023" "01-701-1028" "01-701-1033" ...
..- attr(*, "label")= chr "Unique Subject Identifier"
..- attr(*, "width")= num 11
$ SUBJID : chr [1:306] "1015" "1023" "1028" "1033" ...
..- attr(*, "label")= chr "Subject Identifier for the Study"
..- attr(*, "width")= num 4
$ RFSTDTC : chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Subject Reference Start Date/Time"
..- attr(*, "width")= num 20
$ RFENDTC : chr [1:306] "2014-07-02" "2012-09-02" "2014-01-14" "2014-04-14" ...
..- attr(*, "label")= chr "Subject Reference End Date/Time"
..- attr(*, "width")= num 20
$ RFXSTDTC: chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Date/Time of First Study Treatment"
..- attr(*, "width")= num 10
$ RFXENDTC: chr [1:306] "2014-07-02" "2012-09-01" "2014-01-14" "2014-03-31" ...
..- attr(*, "label")= chr "Date/Time of Last Study Treatment"
..- attr(*, "width")= num 10
$ RFICDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Informed Consent"
..- attr(*, "width")= num 0
$ RFPENDTC: chr [1:306] "2014-07-02T11:45" "2013-02-18" "2014-01-14T11:10" "2014-09-15" ...
..- attr(*, "label")= chr "Date/Time of End of Participation"
..- attr(*, "width")= num 16
$ DTHDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Death"
..- attr(*, "width")= num 10
$ DTHFL : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Subject Death Flag"
..- attr(*, "width")= num 1
$ SITEID : chr [1:306] "701" "701" "701" "701" ...
..- attr(*, "label")= chr "Study Site Identifier"
..- attr(*, "width")= num 3
$ AGE : num [1:306] 63 64 71 74 77 85 59 68 81 84 ...
..- attr(*, "label")= chr "Age"
..- attr(*, "width")= num 8
$ AGEU : chr [1:306] "YEARS" "YEARS" "YEARS" "YEARS" ...
..- attr(*, "label")= chr "Age Units"
..- attr(*, "width")= num 5
$ SEX : chr [1:306] "F" "M" "M" "M" ...
..- attr(*, "label")= chr "Sex"
..- attr(*, "width")= num 1
$ RACE : chr [1:306] "WHITE" "WHITE" "WHITE" "WHITE" ...
..- attr(*, "label")= chr "Race"
..- attr(*, "width")= num 32
$ ETHNIC : chr [1:306] "HISPANIC OR LATINO" "HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" ...
..- attr(*, "label")= chr "Ethnicity"
..- attr(*, "width")= num 22
$ ARMCD : chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Planned Arm Code"
..- attr(*, "width")= num 8
$ ARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
..- attr(*, "width")= num 20
$ ACTARMCD: chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Actual Arm Code"
..- attr(*, "width")= num 8
$ ACTARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
..- attr(*, "width")= num 20
$ COUNTRY : chr [1:306] "USA" "USA" "USA" "USA" ...
..- attr(*, "label")= chr "Country"
..- attr(*, "width")= num 3
$ DMDTC : chr [1:306] "2013-12-26" "2012-07-22" "2013-07-11" "2014-03-10" ...
..- attr(*, "label")= chr "Date/Time of Collection"
..- attr(*, "width")= num 10
$ DMDY : num [1:306] -7 -14 -8 -8 -7 -21 NA -9 -13 -7 ...
..- attr(*, "label")= chr "Study Day of Collection"
..- attr(*, "width")= num 8
$ TRT01P : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
..- attr(*, "width")= num 20
$ TRT01A : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
..- attr(*, "width")= num 20
$ TRTSDTM : POSIXct[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTSTMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "width")= num 1
$ TRTEDTM : POSIXct[1:306], format: "2014-07-02 23:59:59" "2012-09-01 23:59:59" ...
$ TRTETMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "width")= num 1
$ TRTSDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTEDT : Date[1:306], format: "2014-07-02" "2012-09-01" ...
$ TRTDURD : num [1:306] 182 28 180 14 183 26 NA 190 10 55 ...
..- attr(*, "width")= num 8
$ SCRFDT : Date[1:306], format: NA NA ...
$ EOSDT : Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ EOSSTT : chr [1:306] "COMPLETED" "DISCONTINUED" "COMPLETED" "DISCONTINUED" ...
..- attr(*, "width")= num 12
$ FRVDT : Date[1:306], format: NA "2013-02-18" ...
$ RANDDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ DTHDT : Date[1:306], format: NA NA ...
$ DTHDTF : chr [1:306] NA NA NA NA ...
..- attr(*, "width")= num 0
$ DTHADY : num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "width")= num 8
$ LDDTHELD: num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "width")= num 8
$ LSTALVDT: Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ SAFFL : chr [1:306] "Y" "Y" "Y" "Y" ...
..- attr(*, "width")= num 1
$ RACEGR1 : chr [1:306] "White" "White" "White" "White" ...
..- attr(*, "width")= num 9
$ AGEGR1 : chr [1:306] "18-64" "18-64" ">64" ">64" ...
..- attr(*, "width")= num 5
$ REGION1 : chr [1:306] "NA" "NA" "NA" "NA" ...
..- attr(*, "width")= num 2
$ LDDTHGR1: chr [1:306] NA NA NA NA ...
..- attr(*, "width")= num 5
$ DTH30FL : chr [1:306] NA NA NA NA ...
..- attr(*, "width")= num 1
$ DTHA30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "width")= num 0
$ DTHB30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "width")= num 1
- attr(*, "label")= chr "Demographics"
- attr(*, "_xportr.df_arg_")= chr "ADSL"Just like we did for xportr_type(), setting
verbose = "stop" immediately stops R from processing the
lengths. Here the function detects the missing variables and will not
apply any lengths to the dataset until corrective action is applied.
adsl_length <- xportr_length(
.df = ADSL,
metadata = var_spec,
domain = "ADSL",
verbose = "stop",
length_source = "metadata"
)
── Variable lengths missing from metadata. ──
✔ 30 lengths resolved `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
Error in `xportr_logger()` at xportr/R/messages.R:148:7:
! Variable(s) present in dataframe but doesn't exist in `metadata`.Problem with `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
xportr_label()
As you are creating your dataset in R you will often find that R
removes the label of your variable. Using xportr_label()
you can easily re-apply all your labels to your variables in one quick
action.
For this example, we are going to manipulate both the metadata and
the ADSL dataset:
- The metadata will have the variable
TRTSDTlabel be greater than 40 characters. - The
ADSLdataset will have all its labels stripped from it.
Remember in the length example, the labels were on the original
dataset as seen in the str() output.
var_spec_lbl <- var_spec %>%
mutate(label = if_else(variable == "TRTSDT",
"Length of variable label must be 40 characters or less", label
))
adsl_lbl <- ADSL
adsl_lbl <- haven::zap_label(ADSL)We have successfully removed all the labels.
tibble [306 × 51] (S3: tbl_df/tbl/data.frame)
$ STUDYID : chr [1:306] "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" ...
$ USUBJID : chr [1:306] "01-701-1015" "01-701-1023" "01-701-1028" "01-701-1033" ...
$ SUBJID : chr [1:306] "1015" "1023" "1028" "1033" ...
$ RFSTDTC : chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
$ RFENDTC : chr [1:306] "2014-07-02" "2012-09-02" "2014-01-14" "2014-04-14" ...
$ RFXSTDTC: chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
$ RFXENDTC: chr [1:306] "2014-07-02" "2012-09-01" "2014-01-14" "2014-03-31" ...
$ RFICDTC : chr [1:306] NA NA NA NA ...
$ RFPENDTC: chr [1:306] "2014-07-02T11:45" "2013-02-18" "2014-01-14T11:10" "2014-09-15" ...
$ DTHDTC : chr [1:306] NA NA NA NA ...
$ DTHFL : chr [1:306] NA NA NA NA ...
$ SITEID : chr [1:306] "701" "701" "701" "701" ...
$ AGE : num [1:306] 63 64 71 74 77 85 59 68 81 84 ...
$ AGEU : chr [1:306] "YEARS" "YEARS" "YEARS" "YEARS" ...
$ SEX : chr [1:306] "F" "M" "M" "M" ...
$ RACE : chr [1:306] "WHITE" "WHITE" "WHITE" "WHITE" ...
$ ETHNIC : chr [1:306] "HISPANIC OR LATINO" "HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" ...
$ ARMCD : chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
$ ARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
$ ACTARMCD: chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
$ ACTARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
$ COUNTRY : chr [1:306] "USA" "USA" "USA" "USA" ...
$ DMDTC : chr [1:306] "2013-12-26" "2012-07-22" "2013-07-11" "2014-03-10" ...
$ DMDY : num [1:306] -7 -14 -8 -8 -7 -21 NA -9 -13 -7 ...
$ TRT01P : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
$ TRT01A : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
$ TRTSDTM : POSIXct[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTSTMF : chr [1:306] "H" "H" "H" "H" ...
$ TRTEDTM : POSIXct[1:306], format: "2014-07-02 23:59:59" "2012-09-01 23:59:59" ...
$ TRTETMF : chr [1:306] "H" "H" "H" "H" ...
$ TRTSDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTEDT : Date[1:306], format: "2014-07-02" "2012-09-01" ...
$ TRTDURD : num [1:306] 182 28 180 14 183 26 NA 190 10 55 ...
$ SCRFDT : Date[1:306], format: NA NA ...
$ EOSDT : Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ EOSSTT : chr [1:306] "COMPLETED" "DISCONTINUED" "COMPLETED" "DISCONTINUED" ...
$ FRVDT : Date[1:306], format: NA "2013-02-18" ...
$ RANDDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ DTHDT : Date[1:306], format: NA NA ...
$ DTHDTF : chr [1:306] NA NA NA NA ...
$ DTHADY : num [1:306] NA NA NA NA NA NA NA NA NA NA ...
$ LDDTHELD: num [1:306] NA NA NA NA NA NA NA NA NA NA ...
$ LSTALVDT: Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ SAFFL : chr [1:306] "Y" "Y" "Y" "Y" ...
$ RACEGR1 : chr [1:306] "White" "White" "White" "White" ...
$ AGEGR1 : chr [1:306] "18-64" "18-64" ">64" ">64" ...
$ REGION1 : chr [1:306] "NA" "NA" "NA" "NA" ...
$ LDDTHGR1: chr [1:306] NA NA NA NA ...
$ DTH30FL : chr [1:306] NA NA NA NA ...
$ DTHA30FL: chr [1:306] NA NA NA NA ...
$ DTHB30FL: chr [1:306] NA NA NA NA ...
- attr(*, "label")= chr "Demographics"Using xportr_label() we will apply all the labels from
our metadata to the dataset. Please note again that we are using
verbose = "warn" and the function now reports three types
of validation issues:
- Variables in dataset but not in metadata: The same two issues for
RFXSTDTCandDTHDTCare reported as missing from the metadata file (30 variables total).
- Variables in metadata but not in dataset: An additional check
reports 28 metadata variables that are not found in the dataset.
- Label length validation: An additional message is sent around the
TRTSDTlabel having a length greater than 40.
adsl_lbl <- xportr_label(.df = adsl_lbl, metadata = var_spec_lbl, domain = "ADSL", verbose = "warn")
── Variable labels missing from metadata. ──
✔ 30 labels skipped
Warning: Variable(s) present in dataframe but doesn't exist in `metadata`.
✖ Problem with `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
Warning: Variable(s) present in `metadata` but don't exist in dataframe.
✖ Problem with `SITEGR1`, `TRT01PN`, `TRT01AN`, `AVGDD`, `CUMDOSE`, `AGEGR1N`, `RACEN`, `ITTFL`, `EFFFL`, `COMP8FL`, `COMP16FL`, `COMP24FL`, `DISCONFL`, `DSRAEFL`, `BMIBL`, `BMIBLGR1`, `HEIGHTBL`, `WEIGHTBL`, `EDUCLVL`, `DISONSDT`, `DURDIS`, `DURDSGR1`, `VISIT1DT`, `VISNUMEN`, `RFENDT`, `DCDECOD`, `DCSREAS`, and `MMSETOT`
Warning: Length of variable label must be 40 characters or less.
✖ Problem with `TRTSDT`.Success! All labels have been applied that are present in the both
the metadata and the dataset. However, please note that the
TRTSDT variable has had the label with characters greater
than 40 applied to the dataset and the
RFXSTDTC and DTHDTC have empty variable
labels.
tibble [306 × 51] (S3: tbl_df/tbl/data.frame)
$ STUDYID : chr [1:306] "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" ...
..- attr(*, "label")= chr "Study Identifier"
$ USUBJID : chr [1:306] "01-701-1015" "01-701-1023" "01-701-1028" "01-701-1033" ...
..- attr(*, "label")= chr "Unique Subject Identifier"
$ SUBJID : chr [1:306] "1015" "1023" "1028" "1033" ...
..- attr(*, "label")= chr "Subject Identifier for the Study"
$ RFSTDTC : chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Subject Reference Start Date/Time"
$ RFENDTC : chr [1:306] "2014-07-02" "2012-09-02" "2014-01-14" "2014-04-14" ...
..- attr(*, "label")= chr "Subject Reference End Date/Time"
$ RFXSTDTC: chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr ""
$ RFXENDTC: chr [1:306] "2014-07-02" "2012-09-01" "2014-01-14" "2014-03-31" ...
..- attr(*, "label")= chr ""
$ RFICDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ RFPENDTC: chr [1:306] "2014-07-02T11:45" "2013-02-18" "2014-01-14T11:10" "2014-09-15" ...
..- attr(*, "label")= chr ""
$ DTHDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ DTHFL : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Subject Died?"
$ SITEID : chr [1:306] "701" "701" "701" "701" ...
..- attr(*, "label")= chr "Study Site Identifier"
$ AGE : num [1:306] 63 64 71 74 77 85 59 68 81 84 ...
..- attr(*, "label")= chr "Age"
$ AGEU : chr [1:306] "YEARS" "YEARS" "YEARS" "YEARS" ...
..- attr(*, "label")= chr "Age Units"
$ SEX : chr [1:306] "F" "M" "M" "M" ...
..- attr(*, "label")= chr "Sex"
$ RACE : chr [1:306] "WHITE" "WHITE" "WHITE" "WHITE" ...
..- attr(*, "label")= chr "Race"
$ ETHNIC : chr [1:306] "HISPANIC OR LATINO" "HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" ...
..- attr(*, "label")= chr "Ethnicity"
$ ARMCD : chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr ""
$ ARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
$ ACTARMCD: chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr ""
$ ACTARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr ""
$ COUNTRY : chr [1:306] "USA" "USA" "USA" "USA" ...
..- attr(*, "label")= chr ""
$ DMDTC : chr [1:306] "2013-12-26" "2012-07-22" "2013-07-11" "2014-03-10" ...
..- attr(*, "label")= chr ""
$ DMDY : num [1:306] -7 -14 -8 -8 -7 -21 NA -9 -13 -7 ...
..- attr(*, "label")= chr ""
$ TRT01P : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Planned Treatment for Period 01"
$ TRT01A : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Actual Treatment for Period 01"
$ TRTSDTM : POSIXct[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTSTMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "label")= chr ""
$ TRTEDTM : POSIXct[1:306], format: "2014-07-02 23:59:59" "2012-09-01 23:59:59" ...
$ TRTETMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "label")= chr ""
$ TRTSDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTEDT : Date[1:306], format: "2014-07-02" "2012-09-01" ...
$ TRTDURD : num [1:306] 182 28 180 14 183 26 NA 190 10 55 ...
..- attr(*, "label")= chr "Total Treatment Duration (Days)"
$ SCRFDT : Date[1:306], format: NA NA ...
$ EOSDT : Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ EOSSTT : chr [1:306] "COMPLETED" "DISCONTINUED" "COMPLETED" "DISCONTINUED" ...
..- attr(*, "label")= chr "End of Study Status"
$ FRVDT : Date[1:306], format: NA "2013-02-18" ...
$ RANDDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ DTHDT : Date[1:306], format: NA NA ...
$ DTHDTF : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ DTHADY : num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "label")= chr ""
$ LDDTHELD: num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "label")= chr ""
$ LSTALVDT: Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ SAFFL : chr [1:306] "Y" "Y" "Y" "Y" ...
..- attr(*, "label")= chr "Safety Population Flag"
$ RACEGR1 : chr [1:306] "White" "White" "White" "White" ...
..- attr(*, "label")= chr ""
$ AGEGR1 : chr [1:306] "18-64" "18-64" ">64" ">64" ...
..- attr(*, "label")= chr "Pooled Age Group 1"
$ REGION1 : chr [1:306] "NA" "NA" "NA" "NA" ...
..- attr(*, "label")= chr ""
$ LDDTHGR1: chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ DTH30FL : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ DTHA30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
$ DTHB30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr ""
- attr(*, "label")= chr "Demographics"
- attr(*, "_xportr.df_arg_")= chr "ADSL"Just like we did for the other functions, setting
verbose = "stop" immediately stops R from processing the
labels. Here the function detects the mismatches between the variables
and labels as well as the label that is greater than 40 characters. As
this stops the process, none of the labels will be applied to the
dataset until corrective action is applied.
adsl_label <- xportr_label(.df = adsl_lbl, metadata = var_spec_lbl, domain = "ADSL", verbose = "stop")
── Variable labels missing from metadata. ──
✔ 30 labels skipped
Error in `xportr_logger()` at xportr/R/messages.R:174:5:
! Variable(s) present in dataframe but doesn't exist in `metadata`.
✖ Problem with `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
xportr_order()
The order of the dataset can greatly increase readability of the
dataset for downstream stakeholders. For example, having all the
treatment related variables or analysis variables grouped together can
help with inspection and understanding of the dataset.
xportr_order() can take the order information from the
metadata and apply it to your dataset.
adsl_ord <- xportr_order(.df = ADSL, metadata = var_spec, domain = "ADSL", verbose = "warn")
── 30 variables not in spec and moved to end ──
Warning: Variable moved to end in `.df`: `RFXSTDTC`, `RFXENDTC`, `RFICDTC`,
`RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`,
`DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`,
`RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`,
`REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── 42 reordered in dataset ──
Warning: Variable reordered in `.df`: `SITEID`, `ARM`, `TRT01P`, `TRT01A`,
`TRTSDT`, `TRTEDT`, `TRTDURD`, `AGE`, `AGEGR1`, `AGEU`, `RACE`, `ETHNIC`,
`SAFFL`, `DTHFL`, `RFSTDTC`, `RFENDTC`, `EOSSTT`, `RFXSTDTC`, `RFXENDTC`,
`RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`,
`DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`,
`FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, and
`RACEGR1`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
Warning: Variable(s) present in `metadata` but don't exist in dataframe.
✖ Problem with `SITEGR1`, `TRT01PN`, `TRT01AN`, `AVGDD`, `CUMDOSE`, `AGEGR1N`, `RACEN`, `ITTFL`, `EFFFL`, `COMP8FL`, `COMP16FL`, `COMP24FL`, `DISCONFL`, `DSRAEFL`, `BMIBL`, `BMIBLGR1`, `HEIGHTBL`, `WEIGHTBL`, `EDUCLVL`, `DISONSDT`, `DURDIS`, `DURDSGR1`, `VISIT1DT`, `VISNUMEN`, `RFENDT`, `DCDECOD`, `DCSREAS`, and `MMSETOT`Readers are encouraged to inspect the dataset and metadata to see the
past order and updated order after calling the function. Note the
messaging from xportr_order():
- Variables not in the metadata are moved to the end
- Variables not in order are re-ordered and a message is printed out on which ones were re-ordered.
- Variables in metadata but missing from the dataset are identified and reported
adsl_ord <- xportr_order(.df = ADSL, metadata = var_spec, domain = "ADSL", verbose = "stop")
── 30 variables not in spec and moved to end ──
Error in `xportr_logger()` at xportr/R/messages.R:201:5:
! Variable moved to end in `.df`: `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`Just like we did for the other functions, setting
verbose = "stop" immediately stops R from processing the
order. If variables or metadata are missing from either, the re-ordering
will not process until corrective action is performed.
xportr_format()
Formats play an important role in the SAS language and have a column
in specification files. Being able to easily apply formats into your
xpt file will allow downstream users of SAS to quickly
format the data appropriately when reading into a SAS-based system.
xportr_format() can take these formats and apply them.
Please reference xportr_length() or
xportr_label() to note the missing attr() for
formats in our ADSL dataset.
This example is slightly different from previous examples. You will
need to use xportr_type() to coerce R Date variables and
others types to character or numeric. Only then can you use
xportr_format() to apply the format column to the
dataset.
adsl_fmt <- ADSL %>%
xportr_type(metadata = var_spec, domain = "ADSL", verbose = "warn") %>%
xportr_format(metadata = var_spec, domain = "ADSL")Success! We have taken the metadata formats and applied them to the
dataset. Please inspect variables like TRTSDT or
DISONSDT to see the DATE9. format being
applied.
tibble [306 × 51] (S3: tbl_df/tbl/data.frame)
$ STUDYID : chr [1:306] "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" "CDISCPILOT01" ...
..- attr(*, "label")= chr "Study Identifier"
..- attr(*, "format.sas")= chr ""
$ USUBJID : chr [1:306] "01-701-1015" "01-701-1023" "01-701-1028" "01-701-1033" ...
..- attr(*, "label")= chr "Unique Subject Identifier"
..- attr(*, "format.sas")= chr ""
$ SUBJID : chr [1:306] "1015" "1023" "1028" "1033" ...
..- attr(*, "label")= chr "Subject Identifier for the Study"
..- attr(*, "format.sas")= chr ""
$ RFSTDTC : chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Subject Reference Start Date/Time"
..- attr(*, "format.sas")= chr ""
$ RFENDTC : chr [1:306] "2014-07-02" "2012-09-02" "2014-01-14" "2014-04-14" ...
..- attr(*, "label")= chr "Subject Reference End Date/Time"
..- attr(*, "format.sas")= chr ""
$ RFXSTDTC: chr [1:306] "2014-01-02" "2012-08-05" "2013-07-19" "2014-03-18" ...
..- attr(*, "label")= chr "Date/Time of First Study Treatment"
..- attr(*, "format.sas")= chr ""
$ RFXENDTC: chr [1:306] "2014-07-02" "2012-09-01" "2014-01-14" "2014-03-31" ...
..- attr(*, "label")= chr "Date/Time of Last Study Treatment"
..- attr(*, "format.sas")= chr ""
$ RFICDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Informed Consent"
..- attr(*, "format.sas")= chr ""
$ RFPENDTC: chr [1:306] "2014-07-02T11:45" "2013-02-18" "2014-01-14T11:10" "2014-09-15" ...
..- attr(*, "label")= chr "Date/Time of End of Participation"
..- attr(*, "format.sas")= chr ""
$ DTHDTC : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Date/Time of Death"
..- attr(*, "format.sas")= chr ""
$ DTHFL : chr [1:306] NA NA NA NA ...
..- attr(*, "label")= chr "Subject Death Flag"
..- attr(*, "format.sas")= chr ""
$ SITEID : chr [1:306] "701" "701" "701" "701" ...
..- attr(*, "label")= chr "Study Site Identifier"
..- attr(*, "format.sas")= chr ""
$ AGE : num [1:306] 63 64 71 74 77 85 59 68 81 84 ...
..- attr(*, "label")= chr "Age"
..- attr(*, "format.sas")= chr ""
$ AGEU : chr [1:306] "YEARS" "YEARS" "YEARS" "YEARS" ...
..- attr(*, "label")= chr "Age Units"
..- attr(*, "format.sas")= chr ""
$ SEX : chr [1:306] "F" "M" "M" "M" ...
..- attr(*, "label")= chr "Sex"
..- attr(*, "format.sas")= chr ""
$ RACE : chr [1:306] "WHITE" "WHITE" "WHITE" "WHITE" ...
..- attr(*, "label")= chr "Race"
..- attr(*, "format.sas")= chr ""
$ ETHNIC : chr [1:306] "HISPANIC OR LATINO" "HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" "NOT HISPANIC OR LATINO" ...
..- attr(*, "label")= chr "Ethnicity"
..- attr(*, "format.sas")= chr ""
$ ARMCD : chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Planned Arm Code"
..- attr(*, "format.sas")= chr ""
$ ARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
..- attr(*, "format.sas")= chr ""
$ ACTARMCD: chr [1:306] "Pbo" "Pbo" "Xan_Hi" "Xan_Lo" ...
..- attr(*, "label")= chr "Actual Arm Code"
..- attr(*, "format.sas")= chr ""
$ ACTARM : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
..- attr(*, "format.sas")= chr ""
$ COUNTRY : chr [1:306] "USA" "USA" "USA" "USA" ...
..- attr(*, "label")= chr "Country"
..- attr(*, "format.sas")= chr ""
$ DMDTC : chr [1:306] "2013-12-26" "2012-07-22" "2013-07-11" "2014-03-10" ...
..- attr(*, "label")= chr "Date/Time of Collection"
..- attr(*, "format.sas")= chr ""
$ DMDY : num [1:306] -7 -14 -8 -8 -7 -21 NA -9 -13 -7 ...
..- attr(*, "label")= chr "Study Day of Collection"
..- attr(*, "format.sas")= chr ""
$ TRT01P : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Planned Arm"
..- attr(*, "format.sas")= chr ""
$ TRT01A : chr [1:306] "Placebo" "Placebo" "Xanomeline High Dose" "Xanomeline Low Dose" ...
..- attr(*, "label")= chr "Description of Actual Arm"
..- attr(*, "format.sas")= chr ""
$ TRTSDTM : POSIXct[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTSTMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "format.sas")= chr ""
$ TRTEDTM : POSIXct[1:306], format: "2014-07-02 23:59:59" "2012-09-01 23:59:59" ...
$ TRTETMF : chr [1:306] "H" "H" "H" "H" ...
..- attr(*, "format.sas")= chr ""
$ TRTSDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ TRTEDT : Date[1:306], format: "2014-07-02" "2012-09-01" ...
$ TRTDURD : num [1:306] 182 28 180 14 183 26 NA 190 10 55 ...
..- attr(*, "format.sas")= chr ""
$ SCRFDT : Date[1:306], format: NA NA ...
$ EOSDT : Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ EOSSTT : chr [1:306] "COMPLETED" "DISCONTINUED" "COMPLETED" "DISCONTINUED" ...
..- attr(*, "format.sas")= chr ""
$ FRVDT : Date[1:306], format: NA "2013-02-18" ...
$ RANDDT : Date[1:306], format: "2014-01-02" "2012-08-05" ...
$ DTHDT : Date[1:306], format: NA NA ...
$ DTHDTF : chr [1:306] NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ DTHADY : num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ LDDTHELD: num [1:306] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ LSTALVDT: Date[1:306], format: "2014-07-02" "2012-09-02" ...
$ SAFFL : chr [1:306] "Y" "Y" "Y" "Y" ...
..- attr(*, "format.sas")= chr ""
$ RACEGR1 : chr [1:306] "White" "White" "White" "White" ...
..- attr(*, "format.sas")= chr ""
$ AGEGR1 : chr [1:306] "18-64" "18-64" ">64" ">64" ...
..- attr(*, "format.sas")= chr ""
$ REGION1 : chr [1:306] "NA" "NA" "NA" "NA" ...
..- attr(*, "format.sas")= chr ""
$ LDDTHGR1: chr [1:306] NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ DTH30FL : chr [1:306] NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ DTHA30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
$ DTHB30FL: chr [1:306] NA NA NA NA ...
..- attr(*, "format.sas")= chr ""
- attr(*, "label")= chr "Demographics"
- attr(*, "_xportr.df_arg_")= chr "ADSL"At the time of {xportr} v0.3.0 we have not implemented
any warnings or error messaging for this function. However,
xportr_write() through xpt_validate() will
check that formats applied are valid SAS formats.
xportr_write()
Finally, we want to write out an xpt dataset with all
our metadata applied.
We will make use of xportr_metadata() to reduce
repetitive metadata and domain specifications. We will use default
option for verbose, which is just message and so not set
anything for verbose. In xportr_write() we
will specify the path, which will just be our current working directory,
set the dataset label and toggle the strict_checks to be
FALSE. It is also note worthy that you can set the dataset
label using the xportr_df_label and a
dataset_spec which will be used by the
xportr_write()
ADSL %>%
xportr_metadata(var_spec, "ADSL") %>%
xportr_type() %>%
xportr_length(length_source = "metadata") %>%
xportr_label() %>%
xportr_order() %>%
xportr_format() %>%
xportr_df_label(dataset_spec) %>%
xportr_write(path = "adsl.xpt", strict_checks = FALSE)
── Variable lengths missing from metadata. ──
✔ 30 lengths resolved `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── Variable labels missing from metadata. ──
✔ 30 labels skipped
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── 30 variables not in spec and moved to end ──
── 42 reordered in dataset ──
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skippedSuccess! We have applied types, lengths, labels, ordering and formats
to our dataset. Note the messages written out to the console. Remember
the TRTDUR and DCREASCD and how these are not
present in the metadata, but in the dataset. This impacts the messaging
for lengths and labels where xportr is printing out some
feedback to us on the two issues. 5 types are coerced, as well as 36
variables re-ordered. Note that strict_checks was set to
FALSE.
The next two examples showcase the strict_checks = TRUE
option in xportr_write() where we will look at formats and
labels.
ADSL %>%
xportr_write(path = "adsl.xpt", metadata = dataset_spec, domain = "ADSL", strict_checks = TRUE)As there at several ---DT type variables,
xportr_write() detects the lack of formats being applied.
To correct this remember you can use xportr_type() and
xportr_format() to apply formats to your xpt dataset.
Below we have manipulated the labels to again be greater than 40
characters for TRTSDT. We have turned off
xportr_label() verbose options to only produce a message.
However, xportr_write() with
strict_checks = TRUE will error out as this is one of the
many xpt_validate() checks going on behind the scenes.
var_spec_lbl <- var_spec %>%
mutate(label = if_else(variable == "TRTSDT",
"Length of variable label must be 40 characters or less", label
))
ADSL %>%
xportr_metadata(var_spec_lbl, "ADSL") %>%
xportr_label() %>%
xportr_type() %>%
xportr_format() %>%
xportr_df_label(dataset_spec) %>%
xportr_write(path = "adsl.xpt", strict_checks = TRUE)
── Variable labels missing from metadata. ──
✔ 30 labels skipped
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
Warning: Length of variable label must be 40 characters or less.
✖ Problem with `TRTSDT`.
Error in `xportr_write()`:
! The following validation checks failed:
• Label 'TRTSDT=Length of variable label must be 40 characters or less' must be 40 characters or less.
xportr()
Too many functions to call? Simplify with xportr(). It
bundles all core xportr functions for writing to
xpt.
xportr(
ADSL,
var_metadata = var_spec,
df_metadata = dataset_spec,
domain = "ADSL",
verbose = "none",
path = "adsl.xpt"
)
── Variable lengths missing from metadata. ──
✔ 30 lengths resolved `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── Variable labels missing from metadata. ──
✔ 30 labels skipped
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── 30 variables not in spec and moved to end ──
── 42 reordered in dataset ──
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skippedxportr() is equivalent to calling the following
functions individually:
ADSL %>%
xportr_metadata(var_spec, "ADSL") %>%
xportr_type() %>%
xportr_length(length_source = "metadata") %>%
xportr_label() %>%
xportr_order() %>%
xportr_format() %>%
xportr_df_label(dataset_spec) %>%
xportr_write(path = "adsl.xpt", strict_checks = FALSE)
── Variable lengths missing from metadata. ──
✔ 30 lengths resolved `RFXSTDTC`, `RFXENDTC`, `RFICDTC`, `RFPENDTC`, `DTHDTC`, `ARMCD`, `ACTARMCD`, `ACTARM`, `COUNTRY`, `DMDTC`, `DMDY`, `TRTSDTM`, `TRTSTMF`, `TRTEDTM`, `TRTETMF`, `SCRFDT`, `EOSDT`, `FRVDT`, `RANDDT`, `DTHDT`, `DTHDTF`, `DTHADY`, `LDDTHELD`, `LSTALVDT`, `RACEGR1`, `REGION1`, `LDDTHGR1`, `DTH30FL`, `DTHA30FL`, and `DTHB30FL`
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── Variable labels missing from metadata. ──
✔ 30 labels skipped
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skipped
── 30 variables not in spec and moved to end ──
── 42 reordered in dataset ──
── Variables in metadata not found in dataset. ──
✔ 28 metadata variables skippedFuture Work
xportr is still undergoing development. We hope to
produce more vignettes and functions that will allow users to bulk
process multiple datasets as well as have examples of piping
xpt files and related documentation to a validation
software service. As always, please let us know of any feature requests,
documentation updates or bugs on our GitHub
repo.
