Workshop Goals
This workshop is not exhaustive but meant to be a first contact with the R programming language. We hope you leave the workshop able to say:
R isn’t scary!
We also hope to show you you can use R to do things with data you’re already familiar with, as well as clinical computations.
Object Types
Data Frames in R are like Datasets in SAS®. Data frames are made up of columns called vectors – treated like Variables in SAS® More data types exist, but we’ll focus on data frames
Basic Variable Types
Numeric
Character
Boolean
| a | b | c |
|---|---|---|
| 1 | a | TRUE |
| 2 | b | TRUE |
| 3 | c | FALSE |
Assigning Variables
These two methods yield the same results, but the convention is to use <-. Learn more here
Testing Equality
| Operator | Meaning | Example |
|---|---|---|
| <- | assign | x <- y |
| == | equal to | x == y |
| != | not equal to | x != y |
| < | less than | x < y |
| <= | less than or equal to | x <= y |
| > | greater than | x > y |
| >= | greater than or equal to | x >= y |
Arithmetic Operators
| operator | Meaning | Example | Result |
|---|---|---|---|
| + | addition | 1 + 1 == 2 |
2 |
| - | subtraction | 1 -1 == 0 |
0 |
| / | division | 6/3 == 2 |
2 |
| * | multiplication | 2 * 3 == 6 |
6 |
^ or ** |
exponentiation | 3 ** 2 or 3 ^ 2 |
9 |
%% |
modulus | 6%%5 |
1 |
%% |
integer division | 7 %% 2 |
3 |
A Couple More
| Operator | Meaning | Example |
|---|---|---|
| & | and | x & y |
| |
or | x | y |
| ! | not | !x |
| %in% | in | x %in% y |
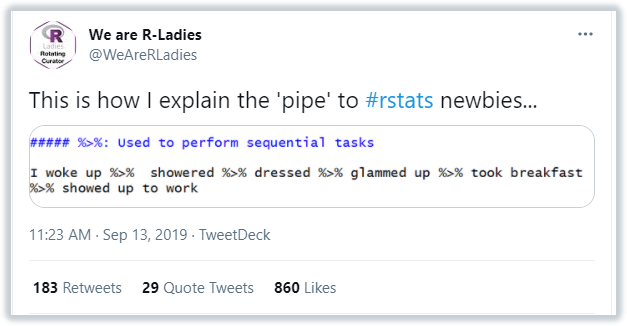
The Pipe %>% Operator
The pipe, %>%, is used to create a pipeline of functions and can be read as “and then”
What are packages?
Packagers are collections of functions and tools to expand the capabilities of R. You can import a package with: library(package_name)
What is the tidyverse?
The tidyverse is an opinionated collection of R packages designed for data science. All packages share an underlying design philosophy, grammar, and data structures.
Install the complete tidyverse with:
install.packages("tidyverse")
Keep:
We can keep only the columns a and b from the original dat:
| a | b | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
With the code:
dat %>%
select(a,b)
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
Drop
We can drop column c, choosing everything but column c:
| a | b | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
By using the code:
dat %>%
select(-c)
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
Sub-setting by rows (where)
We can subset dat where f >= 5
| a | b | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
Using the following code:
dat %>%
filter(b>=5)
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 7 |
Rename
We can use R’s rename function to rename columns a and b to groups and values. Given this starting data frame:
| a | b | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
We can use the code:
dat %>%
rename(
groups = a,
values = b
)
| groups | values | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
Sorting data
Compared to SAS®, you don’t have to sort a lot of the time!
When do I sort? - Presentation - Order dependent operations (i.e. baseline flag) - Don’t need it for grouping
For instance, if we want to sort our data frame by column b:
| a | b | c |
|---|---|---|
| X | 5 | 15 |
| X | 10 | 20 |
| Y | 2 | 12 |
| Y | 7 | 17 |
We can use the arrange function on column b
dat %>%
arrange(b)
| a | b | c |
|---|---|---|
| Y | 2 | 12 |
| X | 5 | 15 |
| Y | 7 | 17 |
| X | 10 | 20 |
set AKA bind_rows
merge AKA *_join
Adding/editing a variable
We can use the mutate function to create new columns using the data from existing columns. For instance we can create a new column c by adding 10 to column b. We can also use the mutate function to or modify existing columns in place. For example, rather than create a new column, we can overwrite column a adding - before and after each entry.
Original Data
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
Using the following code:
dat %>%
mutate(
c = b + 10,
a = paste0("-", a, "-")
)
| a | b | c |
|---|---|---|
| -X- | 5 | 15 |
| -X- | 10 | 20 |
| -Y- | 2 | 12 |
| -Y- | 7 | 17 |
if_else logic
We can use if_else within a mutate to create new columns based on another column. For instance, we can create a categorical column of High and Low values based on column b:
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
We can use this code:
dat %>%
mutate(
level = if_else(b > 5,
"High",
"Low")
)
| a | b | level |
|---|---|---|
| X | 5 | Low |
| X | 10 | High |
| Y | 2 | Low |
| Y | 7 | High |
But what if we want another, Medium category for values greater than 3 but less than 7?
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
If we were to use an if_else statement that would require nesting
dat %>%
mutate(
level = if_else(b < 3, "Low",
if_else(b < 8, "Mid", "High"))
)
But this is really hard to read! Lucky for us we can use the case_when function.
The structure of case_when can be read as:
Left side of
~True/False or something that evaluates to True/FalseRight side of
~Value to return
dat %>%
mutate(
level = case_when(
b < 3 ~ "Low",
b < 8 ~ "Mid",
TRUE ~ "High"
)
)
| a | b | level |
|---|---|---|
| X | 5 | Medium |
| X | 10 | High |
| Y | 2 | Low |
| Y | 7 | Medium |
rowwise vs column operators
Descriptive Statistics
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
| mean | sd | min | max |
|---|---|---|---|
| 6 | 3.36 | 2 | 10 |
Grouped Descriptive Statistics
| a | b |
|---|---|
| X | 5 |
| X | 10 |
| Y | 2 |
| Y | 7 |
| a | mean | sd | min | max |
|---|---|---|---|---|
| X | 7.5 | 3.53 | 5 | 10 |
| Y | 4.5 | 3.53 | 2 | 7 |
Counting
Option 1
| a | b |
|---|---|
| X | dog |
| X | cat |
| X | rabbit |
| Y | rabbit |
| Y | rabbit |
dat %>%
group_by(b) %>%
summarize(
n = n()
)
| a | b |
|---|---|
| 1 | dog |
| 1 | cat |
| 3 | rabbit |
Option 2
| a | b |
|---|---|
| X | dog |
| X | cat |
| X | rabbit |
| Y | rabbit |
| Y | rabbit |
dat %>%
count(b)
| a | b |
|---|---|
| 1 | dog |
| 1 | cat |
| 3 | rabbit |
Grouped Option 1
| a | b |
|---|---|
| X | dog |
| X | cat |
| X | rabbit |
| Y | rabbit |
| Y | rabbit |
dat %>%
group_by(a, b) %>%
summarize(
n = n()
)
| a | b |
|---|---|
| 1 | dog |
| 1 | cat |
| 3 | rabbit |
Other Options
In the course we also showcase 2 other ways to achieve the same goal. The first is using group_by and count
dat %>%
group_by(a) %>%
count(b)
or with even shorter code, calling count on both columns we’d like to group by:
dat %>%
count(a, b)